General about HIV
*Human immunodeficiency virus*
1. Infection spread through blood; semen; vaginal fluid; breast milk as *free virus particles* and as *provirus* within infected cells
2.
Discovered in 1981, and has killed more than 25 million people
3. Listed as a *pandemic* by the WHO
4. Antiretroviral treatment reduces mortality, but only limited access in many countries
5. HIV infects the immunesystem vital cells: Helper T-cell, macrophages and dendritic cells
HIV structure and genome
Retrovirus; enveloped with RNA; about 120 nm in diameter
1. Two copies of sRNA inside a capsid (2,000 copies of *viral protein p24*)
2. RNA bound tightly to nucleocapsid proteins (p7) and enzymes: Reverse transcriptase; proteases and integrase
3.
Matrix (viral protein p17) surrounds the capsid
4. Viral envelope taken from membrane of human cell
5. Envelope contains proteins from the host cell and about 70 copies of HIV protein Env (glycoprotein gp120+gp41)
Retroviral genome (structure)
1. Two copies of the RNA genome (partly base paired)
2. 5'-cap and poly A tail present
3.
RNA complex includes two molecules of cellular tRNA (lys) - a primer for the initiation of reverse transcription
4. Looks like mRNA, but does not serve as mRNA immediately after infection
Retroviral genome (genes)
1. 9 genes, but 15 proteins; GAG, POL and ENV genes ? large polyproteins which are cleaved by virus-encoded protease (GAG and POL) or a host cell protease (ENV)
2. GAG polyprotein: four proteins: matrix; capsid; nucleocapsid; p6
3.
POL polyprotein: protease; reverse transcriptase and integrase
4. ENV polyprotein Gp160: Cleaved by host cell protease to Gp120 and Gp41
Heterogeneity of HIV
Two species:
1. HIV-1: Widespread with high virulence and infectivity
2. HIV-2: Mostly in West Africa
3. High genetic variability is caused by fast replication and high rate of errors (reverse transcriptase is extremely error-prone)
4.
Also, reverse transcriptase can perform recombination if two differen HIV strains infect the same cells
5. Variability or heterogeneity gives resistance to different drugs, thus no effective vaccine
Entry into the cells
1. Mediated by interaction of the virion envelope glycoproteins (gp120) with the CD4 molecule and also with the chemokine corereceptors (CCR5 or CXCR4)
2. Upon binding, gp120 undergoes conformational change that allow binding to the chemokine receptors
3. Different coreceptors for macophage and T-cell -> tropic strains of HIV
HIV infectious cycle
1.
Binding of viral gp120 to CD4 ? interaction with coreceptors ? membrane fusion and entry into cell
2. Reverse transcription
3. Transit of the DNA to the nucleus
4. Integration of viral DNA into random sites in cellular DNA to form provirus
5. Synthesis of viral RNA by cellular RNA p.
II (using provirus template)
6. Synthesis of viral proteins
7. Proteolytic processing of capsid proteins
8. Assembly and budding of virions
Lytic and lysogenic cycle / latency phase
1. Lysogenic cycle: provirus replicates together with cells DNA
2. Latency phase: HIV remains latent until the cell is stimulated during an response to antigen
3.
Lytic cycle: Formation of multiple virions leading to cell death. This formation is controlled by viral regulatory proteins and requires cellular transcription factors expressed upon activiation of the cells (HIV actually kills mostly these cells that currently fight infection)
3. Memory T4 cells: a reservoir of integrated virus that cannot be eliminated
Phases after infection
1. Acute HIV infection: lytic cycle predominates with rapid replication of viruses. Influenza-like symptoms.
T cell count drops (CD8 cells kills infected Tcells)
2. Latency phase (2weeks to 20 years): HIV active in lymphatic organs - dendritic cells. CD4 cells contain provirus
Reverse transcriptase
1. DNA polymerase activity: requires primer with 3'OH; template (RNA/DNA); Lacks-proofreading function (high error rate)
2.
RNase H activity (nuclease specific for RNA:DNA in RNA hybdrids) - in a different domain than polymerase
In vitro reverse transcriptase uses what
Uses oligo-dT: primer complementary to the polyA tail of mRNA
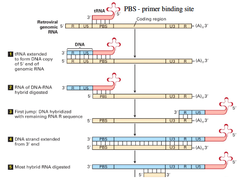
Steps in mechanism in viral reverse transcriptase (first four)
Steps in mechanism of viral reverse trancriptase (last four)
Practical use of reverse transcriptase
1.
Construction of cDNA libraries
2. Detection of specific mRNA:
a. RT-PCR: amplification of cDNA
b. DNA chips (microarrays)
c. Construction of expression vectors containing eukaryotic genes to be expressed in bacterial cells
3.
Monitoring of the anti-HIV therapy effectively - by detection of HIV-RNA in blood (PCR)
How to construct a cDNA library?
1. Using reverse transcriptase with oligo-dT primer. We are able to copy mRNA into cDNA
2. Put this cDNA into e.g. E.
coli which each phage contains a different human cDNA
3. cDNA library reflects the gene expression profile of a given cell type
Expression of microchips (DNA chips)
1. Allow to compare profile of gene expression between two cells
2. Total RNA isolated and cDNA is produced
3. Fluorescently label the cDNA
4.
Equal amounts of each labeled cDNA are applied onto chip and hybridized with probes on the chip
5. Yellow spots respresent equal gene expression in both sample
http://www.genome.gov/10000205 (green dye is the individual)
What is the use of the DNA chip?
Detection of specific sequences
1. Gene polymorphisms
2.
Gene expression analysis
3. Detection of bacterial or viral genomes
HIV testing
1. Initial screening: Enzyme-linked immunosorbent assay (ELISA) - detection of antibodies to HIV-1
2. During the inital phase (2-8 weeks) antibodies may not be detectable.
Infection can be diagnosed by detection of the p24 viral antigen or viral RNA (RT-PCR)
3. Next is confirmatory testing, e.g. western blot
4. Samples which are repeatedly reactive with ELISA and western blot are considered HIV-positive
5.
Real-time PCR can be used to detect number of virions in the blood sample
ELISA mechanism for HIV
1. Partiall purified inactivated HIV antigens pre-coated onto an elisa plate
2. Patient serum is added
3. Anti-human immunoglobulin coupled to an enzyme
4.
Substrate which changes color when cleaved by the enzyme attached to the antibody
(detects antibodies)
Western blot - mechanism
1. HIV lysate proteins are separated by using gel electrophoresis
2. Proteins are blotted onto the membrane
3. Membrane is cut into strips
4. Strips are incubated with patient serum and anti-human IgG conjugated with an enzyme (substrate is added)
5.
The strip is compared with controls
What is important after HIV diagnosis?
1. Monitor the patient viral load
2. HIV viral load - measured by assays which detect HIV-RNA in serum (e.g. RT-PCR)
3.
It is of prognostic value for monitoring the response to antiviral chemotherapy
How does the Real-time PCR work?
1. Amount of PCR amplification products is measured in real time *after each cycle*
2. Allows for the quantitiative analysis of the concentration of the template DNA molecules in the sample
3. The initial number of template molecules is measured based on the number of cycles after which the reaction kientics enters the logarythmic phase
Mechanism of real time PCR
1.
PCR prepared as usual but with the *reporter probe* added (containing fluorescent) between forward and reverse primase
2. Both probe and primers anneal to DNA target
3. Polymerization of new DNA strand is initiated from the primers
4. As the polymerase reaches the probe, its 5'-3' exonuclease degrades the probe
5. This release the fluorescent dye from the quencher and we observe increase in fluoresence
Name some classes of drugs that target HIV during phases of life cycles
1.
Fusion inhibitors (entry inhibitors)
2. Reverse transcriptase inhibitors (NRTI, NNRTI)
3. Ribonuclease inhibitors
4. Intigrase inhibitors
5.
Transcription and translation inhibitors
6. Protease inhibitors
Entry inhibitors
1. Soluble CD4 and antibodies that bind to CD4 antigen
2. *Maraviroc* (approved by FDA in 07), inhibits coreceptor binding
3.
*Fuzeon* or Enfuvirtide; T-20 (approved in 03). Inhibits fusion of membrane by targeting the only known conserved sequence on gp41
Nucleoside reverse transcriptase inhibitors (NRTI)
1. Binds to the enzyme active site
2. 13 Drugs of the type is approved (didanosine, abaclavir, *Azidothymidine AZT*)
Why is azidothymidine (AZT) more important than the others? Side effects and resistance?
1. Higher affinity for reverse transcriptase than for the host cell DNA polymerase
2. It still affects DNA polymerase and can give severe side effects (anemia, neutropenia, myopathy)
3.
Resistance to AZT occurs rapidly when the drug is used in monotherapy
4. It occurs by mutation and makes the affinity less and/or excision of nucleoside analogue from viral DNA by ATP
Non-nucleoside reverse transcriptase inhibitors (NNRTIs)
1. *non-competetive* inhibitors
2. Resistant mutants also emerge rapidly
3. Work against nucleoside analog resistant HIV
4. High affinity / less toxicity
5.
Bind to a *hydrophobic site* on the reverse transcriptase and alters the structure of HIV-1 RT
6. Four drugs approved
Drugs that inhibit integrase activity; why useful
1. Integration of the viral DNA into host cell chromosome is essential for new virus particles
2. Thus, excellent target for specific anti-viral agent - there is no homologous enzyme in humans
3.
*Isentress* was approved in 2007
HIV protease (inhibitors)
1. Dimers which cuts Gag polyprotein into MA,CA,NC
2. Aspartyl protease; high cleave specificty
3. Major class of anti-HIV are protease inhibitors: They are all substrate analogs (peptides) to HIV protease
4. Protease inhibitors can lower the viral load between 1/30 and 1/100 of initial value
What is HAART?
1. Highly active anti-retroviral therapy
2.
Combination of two nucleoside analogs with either protease inhibitor or an NNRTI
3. Viral load should be kept at < 50 copies/ml to prevent emergence of drug-resistant mutants
4. HAART led to major extension of lifespan and suppression of symptoms in HIV-infected patients
5. Not without major side effects (e.
g. abnormal redistribution of body fat (called lipodysptrohy) by protease inhibitors or hemolytic anemia and hemorrhaging (also protease inhibitors))